Garini, Ebenstein, Rabin, Meller, Sivan, Zaslaver, Kafri
 Genomes are dynamic systems undergoing constant changes on a multitude of time scales, ranging from milliseconds to years. These changes are manifested by physical reorganization of the genome within the nucleus (i.e. as a function of cell cycle) and by changes in genomic composition and layout, such as sequence mutations, structural variations, epigenetic chemical modifications and changes in the repertoire and distribution of interacting proteins. This dynamic nature of genomes inherently leads to a high degree of inter- and intra- individual variability on both genetic and epigenetic levels that are greatly responsible for phenotypic variations.
Genomes are dynamic systems undergoing constant changes on a multitude of time scales, ranging from milliseconds to years. These changes are manifested by physical reorganization of the genome within the nucleus (i.e. as a function of cell cycle) and by changes in genomic composition and layout, such as sequence mutations, structural variations, epigenetic chemical modifications and changes in the repertoire and distribution of interacting proteins. This dynamic nature of genomes inherently leads to a high degree of inter- and intra- individual variability on both genetic and epigenetic levels that are greatly responsible for phenotypic variations.
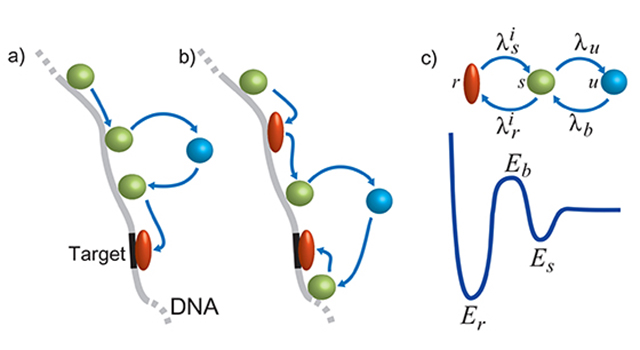
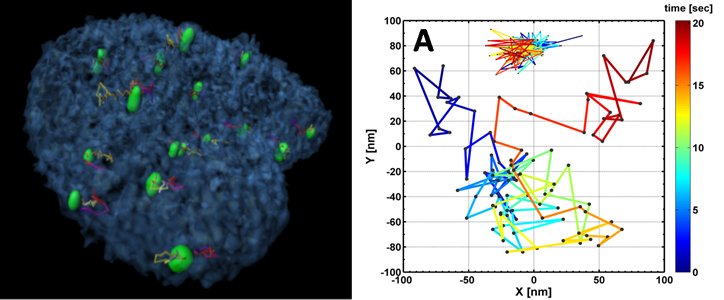
The nucleus forms a crowded environment, containing long DNA molecules that are organized as chromosomes, RNA, proteins, enzymes and a few nuclear bodies. Although the genome is organized in a multi-scale manner, its organizational maintenance mechanisms are not fully understood. Earlier theoretical predictions of chromatin territories and structure of individual DNA molecules have been experimentally confirmed, but it remains unclear how can the interface chromatin both maintain its fractal structure and yet be dynamic enough to allow transcription. The organization was also shown to be important for the proper functioning of the cell. As an example, by observing the telomeres in fixed normal and cancerous cells, it was found that telomeres in normal cells are organized in a cell-cycle dependent manner. Telomere aggregates leads to chromosomal aberrations and c-Myc deregulation leads to the remodeling of the organization. Lately, a study of the architecture of the whole genome with the Hi-C method confirmed the existence of chromosome territories organized in a knot-free fractal globule, as was predicted theoretically. Similar treatment was performed recently to study the properties of polymer layers end-grafted to the inner surface of nanopores connected to solvent reservoirs, as well as for modeling of the ionic conductance of solid-state nanopores, with and without inserted DNA molecule.
The investigation of organizational control mechanisms requires advanced spatio-temporal live-cell imaging methods combined with the development of appropriate biophysical models. In addition, few of the fundamental questions regarding the molecular infrastructure of the chromatin structure and function can efficiently be tested by SM methods such as tethered particle motion (TPM), DNA genotyping and TF interactions using nanopores, optical imaging of stretched chromosomal fragments and force spectroscopy using an AFM. These techniques will be applied to decipher, for example, the role of Lamin A in forming DNA loops in the nucleus.
[/jcolumns] [jcolumns topbordercss=”1px solid #dddddd”] Thermodynamic and dynamic aspects of transcription regulationKaplan, Kafri, Itzkovitz, Haas, Kalisky
Most of current understanding regarding cellular processes leading to transcription ignores the complex circumstances under which transcription occurs, for example: 1) When a Transcription Factor (TF) scans the DNA it experiences a disorder energy landscape, bound to affect the dynamics leading to its binding properties; 2) The non-trivial organization of the DNA, bound to proteins, such as nucleosomes; 3) TFs interaction during scanning of and binding to the DNA; 4) During transcription, RNA polymerase dynamics is significantly affected by secondary structures of the DNA and RNA involved. The latter cardinally affect reverse transcription; 5) Accumulating evidence suggests that the punctilious nature of transcription resonates between open and closed promoters configuration switch, resulting in intermittent bursts of nascent transcripts, shaping the stochastic fluctuations observed in the mRNA content of cells. All of these parameters must be accounted for in order to glean a deeper understanding of transcription. Resolving the multi- step gene expression process in cells and heterogeneous mammalian tissues remains an open challenge.
Using SM techniques it is now feasible to experimentally track TFs as they scan, target and bind to DNA, obtaining a quantitative energy landscape value. Hence, it is currently hypothesized that the energy landscape leads to generic TF classes, each calling for different DNA scanning strategies. Moreover, it is now possible to detect single mRNA molecules in intact cells and even tissues.
[/jcolumns] [jcolumns topbordercss=”1px solid #dddddd”] Translation initiation (TI) dynamics in vitro and in cellsMeller, Haas, Sivan, Garini, Rabin, Levenberg
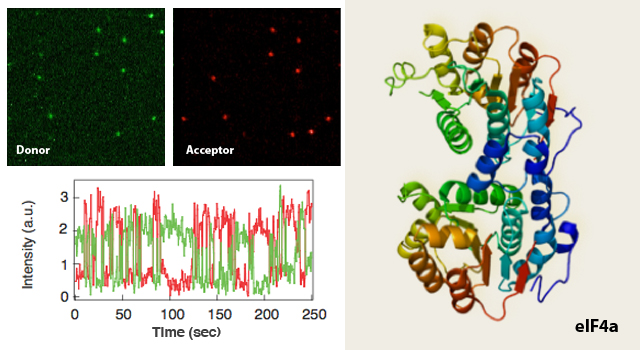 Translation of most mRNAs in eukaryotic cells is regulated at the initiation step, leading to the assembly of the ribosome from the 40S and 60S subunits. The limiting step in this process is the recruitment of the 43S ribosome pre-initiation complex to the 5′-end of the mRNA, marked by an inverted m7GpppN cap. Association of the 43S ribosome complex with the cap is mediated by the cap-binding heterotrimeric complex, eIF4F. The multi-subunit ribosome-bound complex eIF3 and mRNA binding proteins, such as the poly(A) binding protein (PABP) are also associated with eIF4F and are hence involved in TI. Our understanding of TI molecular machinery has progressed tremendously in the past two decades, and we now have a clear view of the multiple components (i.e. factors, kinases, etc.) involved in each stage of the process.
Translation of most mRNAs in eukaryotic cells is regulated at the initiation step, leading to the assembly of the ribosome from the 40S and 60S subunits. The limiting step in this process is the recruitment of the 43S ribosome pre-initiation complex to the 5′-end of the mRNA, marked by an inverted m7GpppN cap. Association of the 43S ribosome complex with the cap is mediated by the cap-binding heterotrimeric complex, eIF4F. The multi-subunit ribosome-bound complex eIF3 and mRNA binding proteins, such as the poly(A) binding protein (PABP) are also associated with eIF4F and are hence involved in TI. Our understanding of TI molecular machinery has progressed tremendously in the past two decades, and we now have a clear view of the multiple components (i.e. factors, kinases, etc.) involved in each stage of the process.
Our longer term goal is to delineate the dynamical details of the entire TI system, in turn applying this knowledge to elucidate its relationship with cancer development and progression. Thus, a detailed description of the dynamical biomolecular processes orchestrating TI will be required, including time scales and forces that govern the association, catalysis and dissociation stages of each step. SM approaches can provide direct, quantitative observations of biomolecular processes and are therefore well suited to address this challenge. We will first focus on characterizing the time-scales associated with eIF4F formation and its interaction with PABP. Fluorescence co-localization and sp-FRET will be employed to quantify RNA unwinding rates of eIF4A, a core component of the 4F complex, responsible for its helicase activity. Subsequently, we will establish 4A’s function in presence of other TI factors, such as the scaffolding protein 4G, the cap-binding 4E and with 4B and 4H – two known to be over-expressed factors in human cancers. Nanopore measurements will enable quantification of the binding forces of the PABP units to the 3’ poly(A) streak of a mRNA. We aim to resolve the question of PABP units cooperative binding onto mRNA and its subsequent impact on the helicase activity. AFM studies will reveal the preferred complex formation along the mRNA structure and assist in the visualization of 4A’s helicase activity.
Recent results published by our collaborators indicate that the activity of the TI apparatus is under tight homeostatic control, operating by regulation of the eIF4E repressor protein 4E-BP1 following ubiquitination. The eIF4F complex serves indeed as the rate-limiting step of TI, rather than the PABP-4E interaction, though the activity of PABP-4E is also homeostatically regulated by the PABP inhibitor, Paip2. The interplay between these two pathways and their biological significance is yet to be uncovered. Therefore, our sp-FRET and NFS studies will be employed to shed light on these multi-complex interactions.
[/jcolumns] [jcolumns topbordercss=”1px solid #dddddd” bottombordercss=”1px solid #dddddd”] Cell differentiation and cell-cell interactions in vitro and in cellsLevenberg, Friedman, Itzkovitz, Zaslaver
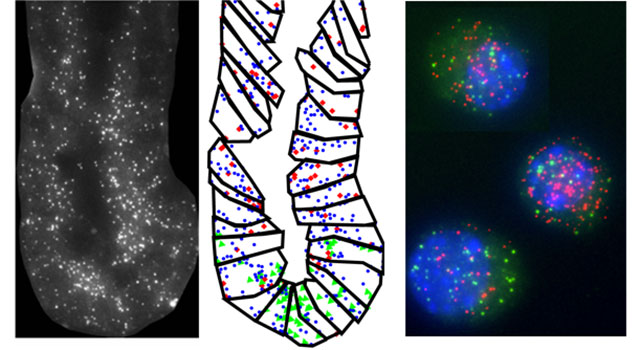 Multicellular organisms are composed of different cell types, which are often renewed throughout life by differentiation of progenitor and stem cells. We view cell differentiation as a dynamic multicellular process, hence aim to investigate roles and mechanisms of intercellular interactions, such as gene expression. We mean to develop a variety of advanced single-cell methods essential for studying heterogeneous cell populations, include, for example, cells at different differentiation stages or mixed phenotypes cells. Also of interest are stochastic processes, such as gene expression and random DNA rearrangement during T cell receptor generation – all deciphered using single-cell studies, baring cardinal affects on cell differentiation.
Multicellular organisms are composed of different cell types, which are often renewed throughout life by differentiation of progenitor and stem cells. We view cell differentiation as a dynamic multicellular process, hence aim to investigate roles and mechanisms of intercellular interactions, such as gene expression. We mean to develop a variety of advanced single-cell methods essential for studying heterogeneous cell populations, include, for example, cells at different differentiation stages or mixed phenotypes cells. Also of interest are stochastic processes, such as gene expression and random DNA rearrangement during T cell receptor generation – all deciphered using single-cell studies, baring cardinal affects on cell differentiation.
The Levenberg group investigates tissue vascularization dynamics both in vitro, using engineered tissues, and in vivo, following implanted tissue constructs integration. Vascular patterning involves signaling between neighboring endothelial cells, which in turn define the rapid switch between leading tip cells and stalk cells, as well as signaling by other cell types in the tissue, together defining the overall pattern. Our aim is to understand how cell migration, gene expression and cellular forces are coperatively coordinated to achieve defined vascular patterns within a defined tissue environment, and how these networks signal the surrounding tissue to affect its differentiation, survival and function. Subsequently, we’ve developed a multicellular model of vascular network formation within engineered muscle (cardiac and skeletal) and pancreatic tissues. Since implanted tissues induce a rapid vascularization process involving anastomosis and perfusion of preexisting tubes with newly formed sprouts, we have designed in vitro and in vivo single/double cell cultures in microfluidic droplets and static adherence array methods to study these cellular interactions. These systems allow a higher resolution response to defined stimulus, thus providing insights into stem cell differentiation and endothelial patterning.
Friedman’s group focuses on differentiation and stochasticity in the immune system, specifically of T lymphocytes. CD4+ T cells can differentiate to at least four different lineages, thereby conferring immune responses against different pathogens. Unbalanced CD4+ T cell differentiation is associated with pathologies such as autoimmunity, allergy and asthma. We study the differentiation process at the single-cell level using live-cell imaging in microfluidics devices. Elevated gene expression stochasticity is observed for cytokines – small, secreted proteins, driving the differentiation process – and cytokine receptors. We investigate the role of gene expression stochasticity while monitoring the expression of these genes during differentiation, looking also at the interplay between stochasticity and intercellular communication. We hypothesize that intercellular interactions can decrease variability in the cell population. We also study the random process of DNA rearrangement that leads to creation of a diverse, though biased, T cell receptor (TCR) repertoire. Use of high-throughput sequencing now allows for comprehensive mapping of these repertoires. We develop methodologies for sequencing and analysis of TCR repertoires, and are interested in understanding general organizing principles of the repertoire and its changes during immune pathologies.
The Itzkovitz lab is combining mathematical models and novel single molecule measurements to study the design principles of stem cell maintained tissues. Our model system is the mouse intestine, a tissue in which long-lived stem cells residing in deep structures called crypts constantly divide to maintain their numbers while producing diverse types of shorter lived cells . We are studying the transcriptional networks that orchestrate the dynamic process of stem cell differentiation in this tissue. By tracing the progenies of individual stem cells and measuring the gene expression signature of individual cells within the intact tissue we aim to achieve a genetic blueprint of this complex differentiation process and to understand the mechanisms that ensure it is carried out robustly.
[/jcolumns]
[jcolumns model=”2,1″]
Participating Universities:
[jcol/]
Funded By:
[/jcolumns][jcolumns model=”2,1″]





[jcol/]


[/jcolumns]